Command Line Reference
The kelp-o-matic package includes one command line tool, kom. It will be registered
in the same Conda environment
that the kelp-o-matic package is installed to.
$ kom --help
Usage: python -m kelp_o_matic.cli [OPTIONS] COMMAND [ARGS]...
Options
--version -v
--install-completion [bash|zsh|fish|powershell|pwsh] Install completion for the specified shell. [default: None]
--show-completion [bash|zsh|fish|powershell|pwsh] Show completion for the specified shell, to copy it or customize the installation. [default: None]
--help -h Show this message and exit.
Commands
find-kelp Detect kelp in image at path SOURCE and output the resulting classification raster to file at path DEST.
find-mussels Detect mussels in image at path SOURCE and output the resulting classification raster to file at path DEST.
find-kelp
$ kom find-kelp --help
Usage: python -m kelp_o_matic.cli find-kelp [OPTIONS] SOURCE DEST
Detect kelp in image at path SOURCE and output the resulting classification raster to file at path DEST.
Arguments
* source TEXT Input image with Byte data type. [default: None] [required]
* dest TEXT File path location to save output to. [default: None] [required]
Options
--species --presence Segment to species or presence/absence level. [default: presence]
--crop-size INTEGER The data window size to run through the segmentation model. [default: 1024]
--rgbi --rgb Use RGB and NIR bands for classification. Assumes RGBI ordering. [default: rgb]
-b INTEGER GDAL-style band re-ordering flag. Defaults to RGB or RGBI order. To e.g., reorder a BGRI image at runtime, pass flags `-b 3 -b 2 -b 1 -b 4`. [default: None]
--gpu --no-gpu Enable or disable GPU, if available. [default: gpu]
--tta --no-tta Use test time augmentation to improve accuracy at the cost of processing time. [default: no-tta]
--help -h Show this message and exit.
Example
Classify kelp species in an RGB image:
kom find-kelp --species --crop-size=1024 ./some/image_with_kelp.tif ./some/place_to_write_output.tif
Classify kelp presence/absence in an BGRI image:
kom find-kelp --rgbi -b 3 -b 2 -b 1 -b 4 --crop-size=1024 ./some/image_with_kelp.tif ./some/place_to_write_output.tif
Tip: Reduce windowed processing artifacts
To reduce artifacts caused by Kelp-O-Matic's moving window classification, use the largest crop_size that you can.
Try starting with a crop_size around 3200 pixels and reduce it if your computer is unable to load that much data at once and the application crashes.
We hope to improve Kelp-O-Matic in the future such that the maximum crop size for your computer can be determined automatically.
CLI Example:
kom --crop-size=3200 your-input-image.tif output-image.tif
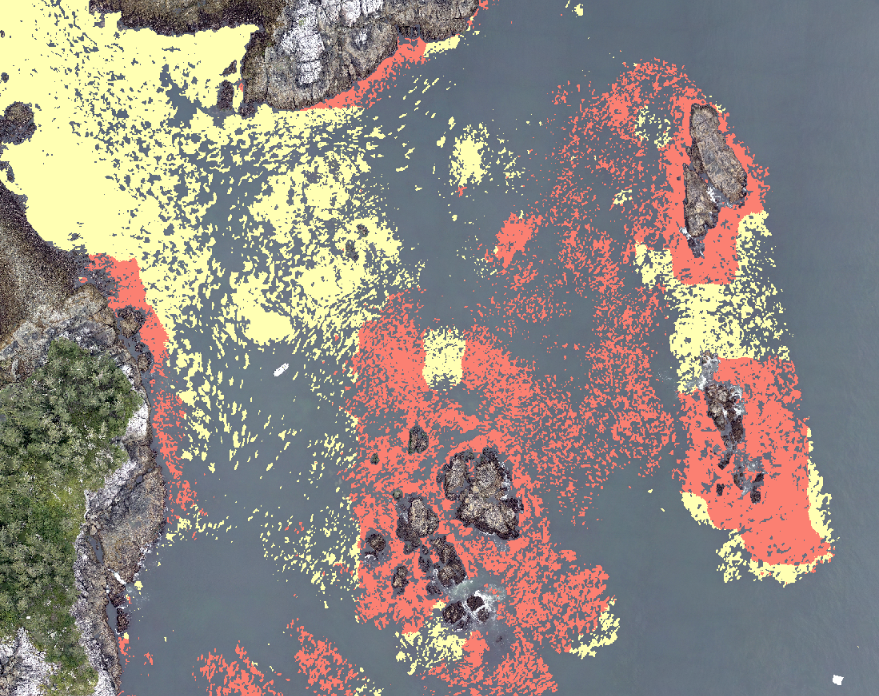
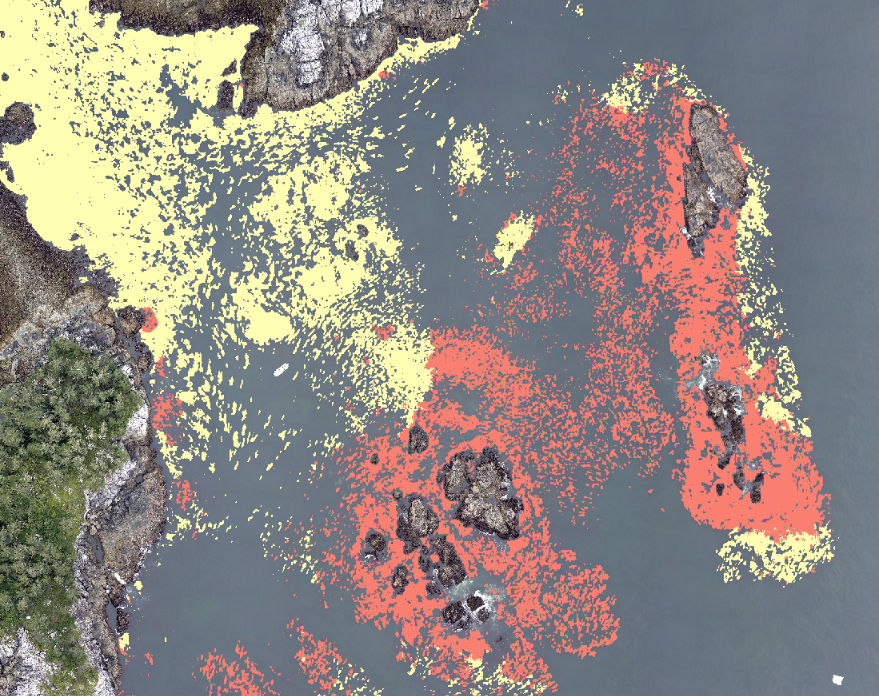
Also worth noting is that the crop_size parameter will change the outputs.
If you need to reproduce a result, make sure to use the same crop_size as the original run.
find-mussels
$ kom find-mussels --help
Usage: python -m kelp_o_matic.cli find-mussels [OPTIONS] SOURCE DEST
Detect mussels in image at path SOURCE and output the resulting classification raster to file at path DEST.
Arguments
* source TEXT Input image with Byte data type. [default: None] [required]
* dest TEXT File path location to save output to. [default: None] [required]
Options
--crop-size INTEGER The data window size to run through the segmentation model. [default: 1024]
-b INTEGER GDAL-style band re-ordering flag. Defaults to RGB order. To e.g., reorder a BGR image at runtime, pass flags `-b 3 -b 2 -b 1`. [default: None]
--gpu --no-gpu Enable or disable GPU, if available. [default: gpu]
--tta --no-tta Use test time augmentation to improve accuracy at the cost of processing time. [default: no-tta]
--help -h Show this message and exit.
Example
kom find-mussels --crop-size=1024 ./some/image_with_mussels.tif ./some/place_to_write_output.tif